


FINAL DIAGNOSIS:
The to 1507del6 variant in the UBE3A gene is previously unreported and of the type expected to cause Angelman Syndrome (See discussion).
DISCUSSION:
Angelman syndrome is characterized by mental retardation, speech impairment, motor dysfunction, movement/balance disorder, and inappropriate laughter. Most patients present with microcephaly, delay in developmental milestones and absence of speech during the first year of life. One of the cardinal behavioral features of Angelman syndrome is a happy disposition with unprovoked laughter, hence the name "Happy Puppet Syndrome". Patients also demonstrate hypopigmentation with fair skin and blue eyes. While our 2-year-old patient has clinical features characteristic for Angelman syndrome, evidence of a genetic defect should be taken into consideration to establish a definitive diagnosis.
Mechanisms of Angelman Syndrome
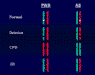
Angelman syndrome is caused by deficiency of gene expression from the maternally inherited chromosome 15q11-q13 region, which is subjected to genetic imprinting. Four classes of genetic mechanisms have been described: maternal deletion, paternal uniparental disomy (UPD), imprinting defects (ID), and mutations within the UBE3A gene (Figures 1 and 2).
The most common mechanism, which occurs in 70-75% of patients, is a large interstitial deletion of ~4Mb on chromosome 15q11-q13 (Figure 1). The normal FISH analysis in this patient rules out the possibility of a large deletion. On the other hand, extensive differential methylation is present across the chromosome 15q11-q13 region. It is most prominent in a CpG island at the 5' end of the SNRPN locus where the maternal chromosome is methylated with SNRPN silenced while the paternal chromosome is unmethylated with SNRPN expressed. In the current case, the methylation analysis of the SNRPN gene showed normal biparental inheritance (Figure 3). These studies cumulatively rule out three of the four classes of Angelman syndrome, i.e., deletion, uniparental disomy, or imprinting center mutation (Figure 1). However, the fourth class, a mutation in the Angelman syndrome gene, UBE3A, cannot be ruled out by the normal SNRPN methylation pattern (Figure 2).
Mutations in UBE3A Gene
The UBE3A gene encodes E6AP-E3 ubiquitin-protein ligase and maps within the 15q11-q13 region on chromosome 15. It is imprinted with paternal silencing specifically in the brain. Loss-of-function mutations of the UBE3A gene constitute 14-38% of sporadic Angelman syndrome cases. Most published mutations are protein truncating and cluster in exons 9 and 16 (Figure 4).
The 1507del6 variant in exon 9 in the current case resulted in the deletion of two amino acids, lysine and valine, at positions 503 and 504. It occurs in a functionally important and highly conserved region of the UBE3A gene and is likely the cause of this patient's disease phenotype.
Interpretation of Sequence Variation and Genetic Counseling
The detected sequence variant is previously unreported. When a novel variant is found, the Molecular Diagnostics report needs to address the likelihood that the sequence change will affect protein expression or function, i.e. whether it is likely to be a mutation associated with the clinical phenotype, a benign polymorphism, or uncertain as to effect. This can have significant impact on genetic counseling.
According to the recent recommendations for standards for interpretation of sequence variations from the American College of Medical Genetics (ACMG), there are five categories of sequence variations for the purposes of clinical reporting: 1) Sequence variation is previously reported and is a recognized cause of the disorder; 2) Sequence variation is previously unreported and is of the type which is expected to cause the disorder; 3) Sequence variation is previously unreported and is of the type which may or may not be causative of the disorder; 4) Sequence variation is previously unreported and is probably not causative of disease; 5) Sequence variation is previously reported and is recognized neutral variant.
In this case, testing the parents for the presence of the variant was helpful. If the father had the same variant and did not display an affected phenotype, it would suggest that the sequence change is a benign polymorphism. However if the mother was a carrier of the sequence variant, a deleterious mutation could not be ruled out. Neither the mother nor the father was a carrier of the 1507del6 variant, suggesting that it is a de novo variant in this patient. The location and type of the sequence change can be very important in predicting the impact of the change in the protein. Changes which result in truncation of the normal protein product through direct introduction of a STOP signal (nonsense mutation) or a shift in the mRNA reading frame (frameshift mutation) almost always are associated with disease. Changes which introduce minor changes in the protein's amino acid sequence, usually substitution of one amino acid for another or sometimes the loss or gain of several amino acids, may also be important if they affect regions that impact protein function such as important functional domains or conserved regions. Finally, nucleotide changes which alter important sequences for protein expression or processing, particularly in upstream regulartory promoter areas or at exon-intron boundaries are often disease-causing.
The sequence variation in the current case is not predicted to shift the mRNA reading frame or result in the introduction of a stop codon, therefore it is not definitively expected to cause the disorder. However, the combination of a milder but typical clinical presentation with the deletion of two amino acids in a highly conserved and functionally important region suggests there is a reasonable likelihood this sequence variant is causative of the patient's phenotype. Conservatively, this sequence variation should be entered in locus-specific database. Follow-up research studies to ascertain clinical significance as well as functional analysis to prove there is compromise of protein function would be useful.
On the other hand, somatic and germline mosaicism for UBE3A mutations has been observed in some mothers of affected children. Although the test results did not show evidence of mosaicism in the mother, they do not exclude its presence. It has been previously reported that, even if the mother tests negative for a mutation identified in a child, there is still an appreciable risk of recurrence due to gonadal mosaicism. Therefore, genetic counseling and prenatal testing should be offered.
SUMMARY:
In summary, this 2-year-old boy with postnatal microcephaly and global developmental delay has clinical features of Angelman syndrome with mild to moderate symptoms. Although diagnostic studies failed to detect the three common defects on chromosome 15 that are responsible for Angelman syndrome, i.e., maternal deletion, paternal uniparental disomy and imprinting defects, sequence analysis revealed a novel sequence variant in exon 9 of UBE3A gene. Laboratory diagnosis and genetic counseling for Angelman syndrome may be complex, and mutation studies of UBE3A may provide useful additional information in patients with normal DNA methylation analysis.
Acknowledgment: Molecular analysis for SNRPN and UBE3A genes was performed by the Department of Genetics at the University of Chicago Medical Center.
REFERENCES:
![]() Contributed by Jing Yu, MD, PhD, Federico Monzon, MD, PhD, Xiao-Ming Yin, MD, PhD, Jeffrey Kant, MD, PhD
Contributed by Jing Yu, MD, PhD, Federico Monzon, MD, PhD, Xiao-Ming Yin, MD, PhD, Jeffrey Kant, MD, PhD