Magee Equations™ for Estimating Oncotype DX® Recurrence Score
Magee Equations™ Input and Results
Insert values in their respective fields. Once all necessary fields have valid values, all three Magee Equations will automatically calculate.|
Nottingham Score 3 to 9 |
ER H-Score 0 to 300 |
PR H-Score 0 to 300 |
HER2 |
Tumor Size cm |
KI-67 Percentage % |
|
Equation 1 Result |
Equation 2 Result |
Equation 3 Result |
|||
| Average Magee Equation Score |
|
||||
Magee Decision Algorithm™
The Magee Decision Algorithmô helps in triaging cases for molecular testing. Use Magee Equation results and mitotic activity score (part of tumor grading) to triage cases.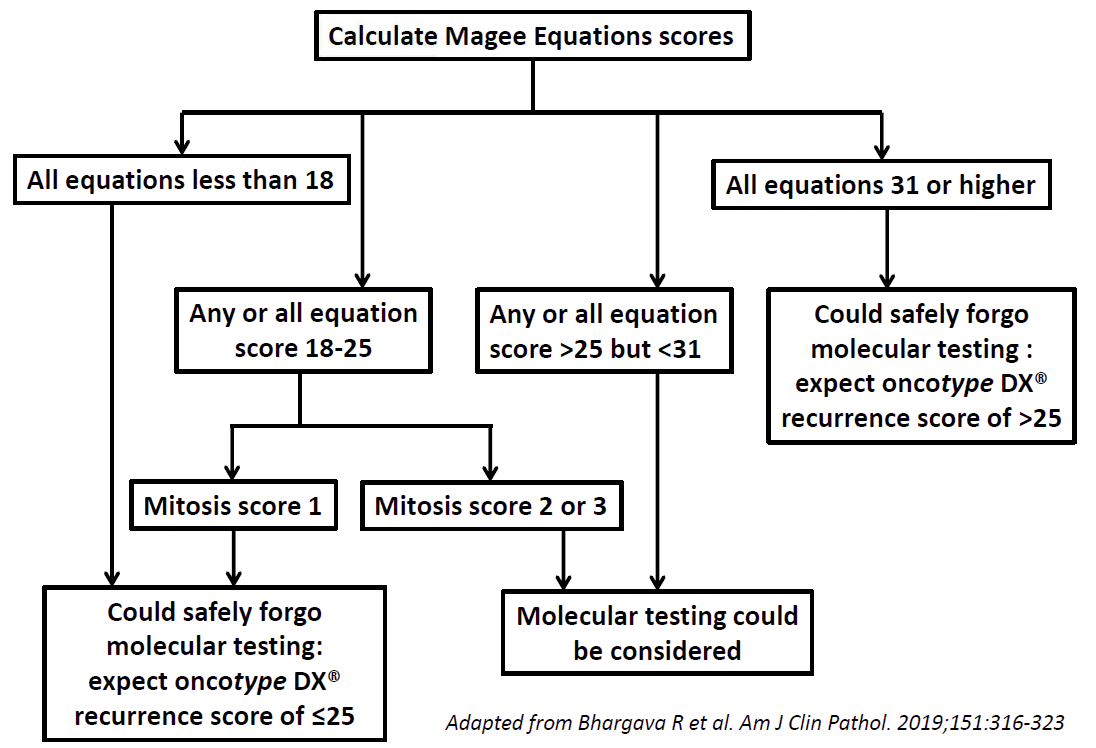
Background
Oncotype DX® (Genomic Health Inc. Redwood City, CA), is a commercial assay, frequently used by oncologists for making chemotherapy decisions. It is a quantitative reverse transcription polymerase chain reaction based assay, used to estimate the risk of distant recurrence for patients with ER positive, lymph node negative breast cancers. It is reported as a numerical score (recurrence score or RS) ranging from 0-100. At test inception, the risk categories were defined as low risk (LR; <18), intermediate risk (IR; 18-30), and high risk (HR; ≥31). In the TAILORx trial, endocrine therapy was non-inferior to chemoendocrine therapy in the analysis of invasive disease-free survival for patients (age >50 years) with RS of 11-25. In a pilot proof of principle study from this institution, it was reported that standard histopathologic factors and immunohistochemical markers can be used to estimate the recurrence score (original Magee equation; Mod Pathol. 2008;21:1255-1261). Subsequently, new Magee Equations were derived using a much larger database of over 800 cases that were sent for clinical oncotype DX® testing to Genomic Health as per oncologistsí request. These new Magee Equations were validated on a separate set of over 200 cases, again sent for clinical oncotype DX® testing to Genomic Health as per oncologistsí request (new Magee Equations; Mod Pathol. 2013;26:658-664).How the New Magee Equations Were Derived
Multiple linear regression analysis was performed to model the prediction of the oncotype DX® RS by Nottingham Score (range 3-9), Ki-67 labeling index (0-100), tumor size (in cm.), H-scores (range: 0-300) for ER and PR, and HER2 status (negative, equivocal or positive). Three models were built based on different hypotheses and data availability. The first regression model included all available parameters (including Ki-67 index) for prediction of oncotype DX® RS. The second regression model was similar to the first but did not include Ki-67. The third regression model included only semi-quantitative immunohistochemical expression levels for ER, PR, HER2 and Ki-67. These 3 equations are now commonly referred to as the Magee Equations™.How to Use the Equations
All the required data is generally present in the surgical pathology report. The hormone receptor results will require the semi-quantitative H-score. If not reported initially, this can be easily calculated by the case pathologist by reviewing the immunohistochemical (IHC) stain slides. H-scores are calculated based on percentage of positive cells showing none, weak, moderate, or strong staining intensity. The score is given as the sum of the percent staining multiplied by an ordinal value corresponding to the intensity level (0=none, 1=weak, 2=moderate, 3=strong). With four intensity levels, the resulting score ranges from 0 (no staining in the tumor) to 300 (diffuse intense staining). For e.g. a tumor showing no staining in 10% cells, weak staining in 30% cells, moderate staining in 40% cells, and strong staining in 20% cells; the H-score will be calculated as follows: (0x10)+(1x30)+(2x40)+(3x20)=170. HER2 status (for purposes of the Magee Equations) should be classified as shown below.- HER2 negative: HER2 IHC score 0, IHC score 1+, and IHC score 2+ with HER2 copies per cell less than 4 by FISH
- HER2 positive: HER2 IHC score of 3+, and HER2 IHC score of 2+ with HER2 copies per cell of 6 or more by FISH
- HER2 equivocal: HER2 IHC score of 2+ with HER2 copies per cell of 4 to less than 6 by FISH
An algorithmic approach (Magee Decision Algorithm™ as illustrated above) to utilize molecular testing in primary breast cancer is discussed in the following publication: https://www.ncbi.nlm.nih.gov/pubmed/30395177 (Bhargava R, et al. 2019). For further validation of Magee Decision Algorithmô, please see: https://www.ncbi.nlm.nih.gov/pubmed/32203092 (Bhargava R, et al. 2020).
Neoadjuvant Chemotherapy and Magee Equation 3 (ME3)
The pathologic complete response rate (pCR) in ER+/HER2-negative tumors increases with increasing ME3 score. Based on available data (PMID: 28548119 and 32661297), the estimated pCR rate is as follows:
- ME3 score <18: pCR rate of 0%
- ME3 score 18-25: pCR rate of <5%
- ME3 score of >25 but <31: pCR rate of 14%
- ME3 score of 31 or higher: pCR rate of 35 to 40%
References - Internal Publications
- Bhargava R et al (2022): https://www.ncbi.nlm.nih.gov/pubmed/36165933
- Bhargava R et al (2020): https://pubmed.ncbi.nlm.nih.gov/32661297/
- Bhargava R et al (2020): https://www.ncbi.nlm.nih.gov/pubmed/32203092
- Soran A et al (2020): https://www.ncbi.nlm.nih.gov/pubmed/32285033
- Bhargava R et al (2019): https://www.ncbi.nlm.nih.gov/pubmed/30395177
- Dabbs DJ et al (2018): https://www.ncbi.nlm.nih.gov/pubmed/29920746
- Farrugia DJ et al (2017): https://www.ncbi.nlm.nih.gov/pubmed/28548119
- Bhargava R (2017): https://www.ncbi.nlm.nih.gov/pubmed/28393310
- Klein ME et al (2013): https://www.ncbi.nlm.nih.gov/pubmed/23503643
- Flanagan M et al (2008): https://www.ncbi.nlm.nih.gov/pubmed/18360352
References - External Publications
- Li H et al (2022): https://www.ncbi.nlm.nih.gov/pubmed/35775006
- Li F et al (2022): https://www.ncbi.nlm.nih.gov/pubmed/35486808
- Glasgow A et al (2021): https://www.ncbi.nlm.nih.gov/pubmed/32951186
- Slembrouck L et al (2021): https://www.ncbi.nlm.nih.gov/pubmed/33558657
- Remoué A et al (2021): https://www.ncbi.nlm.nih.gov/pubmed/33245987
- Robertson SJ et al (2020): https://www.ncbi.nlm.nih.gov/pubmed/31551182
- De Lima MAG et al (2019): https://www.ncbi.nlm.nih.gov/pubmed/31287198
- Turner BM et al (2019): https://www.ncbi.nlm.nih.gov/pubmed/31199586
- Robertson SJ et al (2019): https://www.ncbi.nlm.nih.gov/pubmed/30672056
- Hou Y et al (2018): https://www.ncbi.nlm.nih.gov/pubmed/30084931
- Sughayer M et al (2018): https://www.ncbi.nlm.nih.gov/pubmed/29691722
- Hou Y et al (2017): https://www.ncbi.nlm.nih.gov/pubmed/28898988
- Hou Y et al (2017): https://www.ncbi.nlm.nih.gov/pubmed/28449064
- Harowicz MR et al (2017): https://www.ncbi.nlm.nih.gov/pubmed/28064383
- Chen YY et al (2016): https://www.ncbi.nlm.nih.gov/pubmed/27595437
- Turner BM et al (2015): https://www.ncbi.nlm.nih.gov/pubmed/25932962