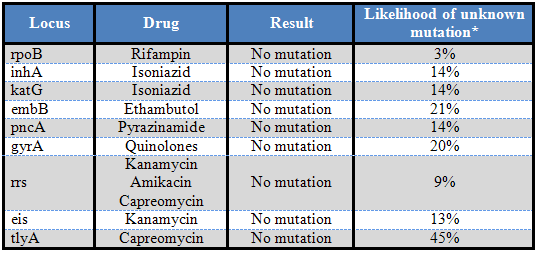
*Based on an examination of 500 isolates at CDC
Sequence Based Susceptibility Results

*Based on an examination of 500 isolates at CDC
All tested loci were negative for mutations. The isolate was also sent for culture based susceptibility testing. Once the patient's liver tests returned to normal, he was gradually re-started on rifampin, isoniazid, ethambutol, and pyrazinamide; he was also placed on moxifloxacin. He had a difficult clinical course, resulting in a repeat admission approximately three weeks after discharge due to poor appetite, weight loss, and fatigue. He was then discharged to short-term rehabilitation for nutritional support.
Subsequent biochemical testing performed on the isolate for niacin and nitrate reduction were positive, indicating that this isolate is indeed Mycobacterium tuberculosis. Phenotypic testing revealed that the patient's isolate was susceptible to rifampin, isoniazid, ethambutol, and pyrazinamide. The Department of Homeland Security indicated that the patient would be required to complete two weeks of effective antituberculous therapy and document three negative AFB smears before he would be permitted to board an airplane.
DISCUSSION
Mycobacterium tuberculosis is still the leading single infectious cause of death throughout the world (1, 2). Within the United States, TB is rare, but has been noted in immigrants and individuals travelling from TB endemic areas. Resistance to anti-tuberculous medications occurs in two predominant forms: MDR (Multi-drug resistant) TB is resistant to two of the primary treatment agents, rifampin and isoniazid (3). XDR (Extensively drug resistant) TB demonstrates resistance at the MDR level, plus resistance to fluoroquinolones (such as ciprofloxacin) and an injectable agent (e.g. aminoglycosides).
In the US, MDR TB is believed to account for approximately 1.1% of new cases (4). However, resistance to single agents is more common, with approximately 8% of new cases resistant to Isoniazid (1), and approximately 1.8% resistant to Rifampin (5). Thus, it is important to assess new cases of tuberculosis for resistance to anti-tuberculous agents. However, due to the slow growing nature of TB, drug sensitivity testing (DST) can take up to 6 weeks to complete depending on the method used (4). This is an inordinate amount of time to wait prior to starting appropriate therapy, so all patients are started on empiric treatment pending the results of sensitivity studies. Molecular drug sensitivity testing is now starting to be available, and can dramatically decrease turn-around times in these cases.
All cases of tuberculosis are reported to the County and State health departments, who also report these cases to the Centers for Disease Control (CDC). There is a quarantine process in effect whereby these patients may be placed on a Do Not Board list (jointly managed by CDC and the Department for Homeland Security), and prevented from obtaining a boarding pass or boarding an aircraft in this country. There are explicit criteria for placement on this list, including if the patient is infectious, non-compliant or unaware of their diagnosis and has expressed intention to travel (6). These lists are accessible by both the airlines and Customs and Border Protection, who may enforce such provisions. In addition, if it is believed that the patient previously traveled while infectious, flight manifests can be requested from the airlines, and the local state and county health departments will attempt to arrange for skin testing for other passengers on the flight. In the case of TB, only passengers sitting in the same row as the index case, as well as passengers two rows in front and two rows behind, will be notified (7).
Genetic mechanisms of drug resistance in TB are complex, but are thought to be primarily due to chromosomal mutations, and are not plasmid mediated (1, 2). These changes occur rather infrequently, with fewer new mutations than would be expected by random variation (3). Given the relatively stable chromosomal mutations that are seen, molecular assays for microbial classification and drug sensitivity have been developed. Many of these tests allow for rapid (within 2 hours) identification of Mycobacterium tuberculosis and determination of preliminary sensitivity patterns. Some of these new tests are even available at the point of care, with minimal technical training required (8). These assays also allow for the separation of Mycobacterium tuberculosis from other atypical mycobacteria, which can be pertinent in guiding empiric therapy (4). Additionally, these tests are more sensitive than traditional techniques and allowed for the detection of AFB smear negative disease in up to 90% of cases in one study (9).
Reference laboratory molecular testing for anti-tubercular drug sensitivity is also available on multiple platforms. Recent studies have demonstrated the viability of the Ion Torrent platform for next generation sequencing, with 100% concordance with phenotypic sensitivity studies, and the ability to characterize mixed strain populations (10, 11). Sanger sequencing is still the gold standard for most of these reference type assays, with focused examination of multiple genetic loci required in order to detect clinically significant mutations. Importantly, the absence of mutation in any of these assays is NOT confirmation of drug susceptibility, as all mechanisms of drug resistance have not yet been elucidated (12). This can cause false negative results if the specific locus is not sequenced. The converse is often true, however, whereby detection of mutation signifies resistance to the selected drug. For these reasons, culture based sensitivity studies still must be performed to ensure that sequence-based testing has not failed to detect an unknown mutation. Current indications for sequence based sensitivity testing include: strains with a high risk of rifampin resistance (based on patient origin or contacts), patients known to have adverse reactions to certain anti-tuberculous drugs, patients where drug resistance will have a high public health impact (such as daycare workers, nurses, and our present patient), mixed or non-viable cultures, and isolates which fail to grow on drug susceptibility testing (12).
Detection of rifampin resistance is most pertinent in many cases due to its usage as a first-line therapeutic agent. Fortunately, the majority (>95%) of rifampin resistant isolates have a single point mutation in an 81bp region of the rpoB gene, enabling this assay to be very sensitive. Mutations in this region alter protein structure so as to prevent rifampin binding (12). Isoniazid resistance is also of concern, as it often occurs concurrently with rifampin resistance (8), and is a primary treatment modality for tuberculosis. There are two important genetic loci examined for isoniazid resistance, including inhA, which encodes the promoter region of the drug target (mutations here lead to overproduction and resultant drug saturation) and katG (mutations at this locus prevent activation of the isoniazid prodrug) (12). Additional genes are also analyzed in many comprehensive molecular sensitivity panels (see above results for entire panel, with likelihood of unknown mutation causing false negative result) (12).
Molecular drug sensitivity testing for tuberculosis provides rapid turnaround with relatively high specificity for the tested agent. Culture based sensitivity must still be completed to ensure genotype-phenotype correlation, but these new assays allow for faster testing, treatment, and cure of this hard to culture and sometimes difficult to treat microorganism.
Acknowledgments
The authors would like to thank Jeffrey Driscoll, PhD at the Centers for Disease Control and Prevention, National Center for Hepatitis, HIV, STD and TB Prevention for his assistance with this case, and subsequent review of this case report.
REFERENCES
![]() Contributed by Ryan A. Collins, MD and A. William Pasculle, ScD
Contributed by Ryan A. Collins, MD and A. William Pasculle, ScD